Towards reproducible science with Jupyter Notebooks

For publications, the scientific principle of reproducibility requires that studies and processes are documented and archived to such a level that they can be repeated at a later point - by the original authors as well as by other interested researchers. Technically, this should be possible for the software-driven steps. However, in practice reproducibility is often not given - for a multitude of reasons.
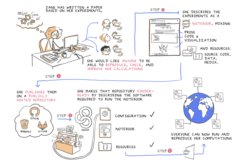
Now researchers from the UK, France, Norway and Germany - led by Hans Fangohr from the MPSD’s new Computational Science unit - have published practical solutions for this challenge on the basis of the OpenDreamKit research project. They recommend reproducible workflows that include the use of open source software and the provision of complex computational capabilities through researcher-friendly high-level command interfaces. These allow scientists to direct the research via electronic notebooks which automatically record every step that has been taken.
The work is published in Computing in Science and Engineering, which has a tradition of publishing important methodological advances.
One of the open source results of this work is the Ubermag simulation environment for magnetism at the nanoscale. The low-level simulation software, that has been optimized for high execution performance, is difficult to use directly, and the researchers developed a high-level Python interface to drive the simulations with much greater ease. The Ubermag environment features visualisation capabilities which provide immediate visual feedback on the simulation results and integrate naturally into the Jupyter notebook. Given this tool set, a simulation study can now be driven from the Jupyter notebook, which combines narrative comments by the researcher with commands to execute the simulation, immediately followed by the results obtained. The notebook has been designed to be able to record and archive this rich and executable document - thus providing a leap forward for the reproducibility of the study.
"To improve reproducibility in science, it is not enough to provide tools or workflows that make the results more reproducible. These tools must also fit into the scientific approach of the researchers and ideally lead to a better research experience, so that the new methods are accepted and adopted widely," says Hans Fangohr, coordinating author of the publication.
Reproducibility is attracting increasing attention from researchers, research councils and publishers. "One additional aspect of the discussion is that reproducibility leads to re-usability,“ Fangohr explains. "If all the steps of a data analysis are documented and archived - ideally in a (potentially long!) computer-executable script - so that they can be repeated automatically at the touch of a button, then it becomes a lot easier to take such a study and extend it. Currently, it is not unusual for PhD students to spend months or years to first reproduce published results as a baseline, before they can extend the work with novel additions as the second step. With the workflows and methods proposed here, we can reduce that time to hours or minutes, making our collective research activity a lot more efficient."